About Insilab
Our activities range from development of purely graph theoretical algorithms to the development of algorithms directly applicable in pharmacy. We continuosly develop the ProBiS web server for prediction of protein binding sites and protein-ligand interactions.
Insilab was created as a repository for open-source software that we developed over the years. It is intended for people who use or develop innovative software for drug discovery.
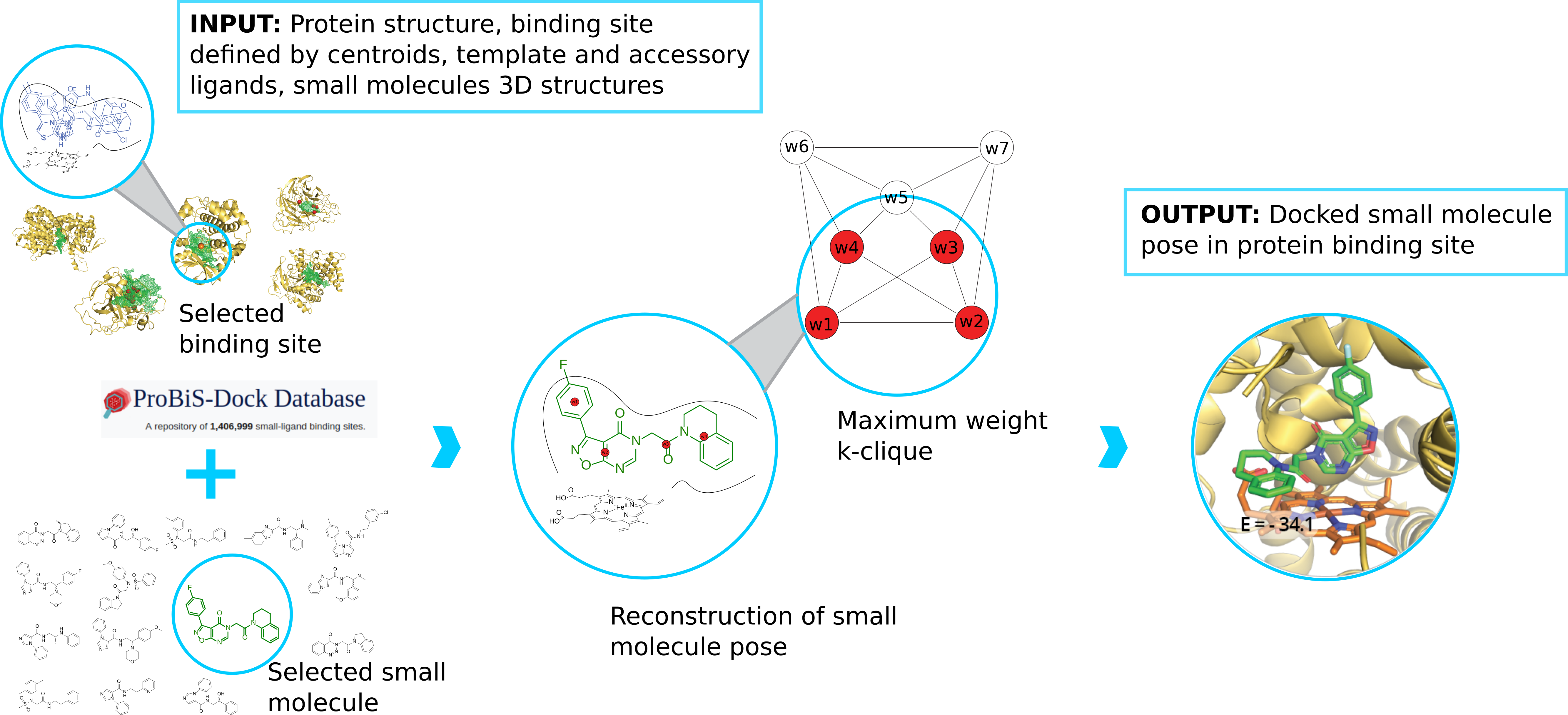
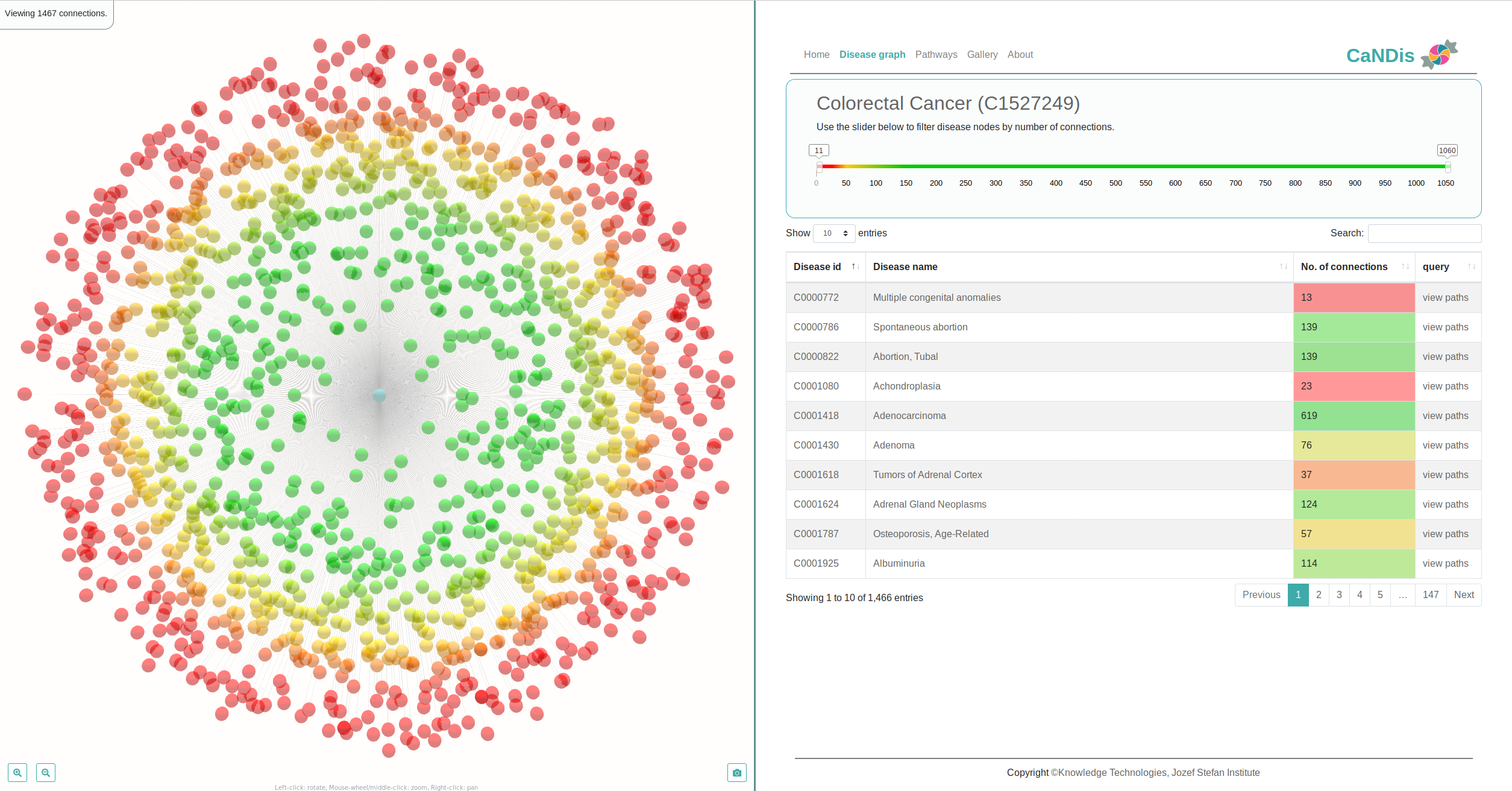
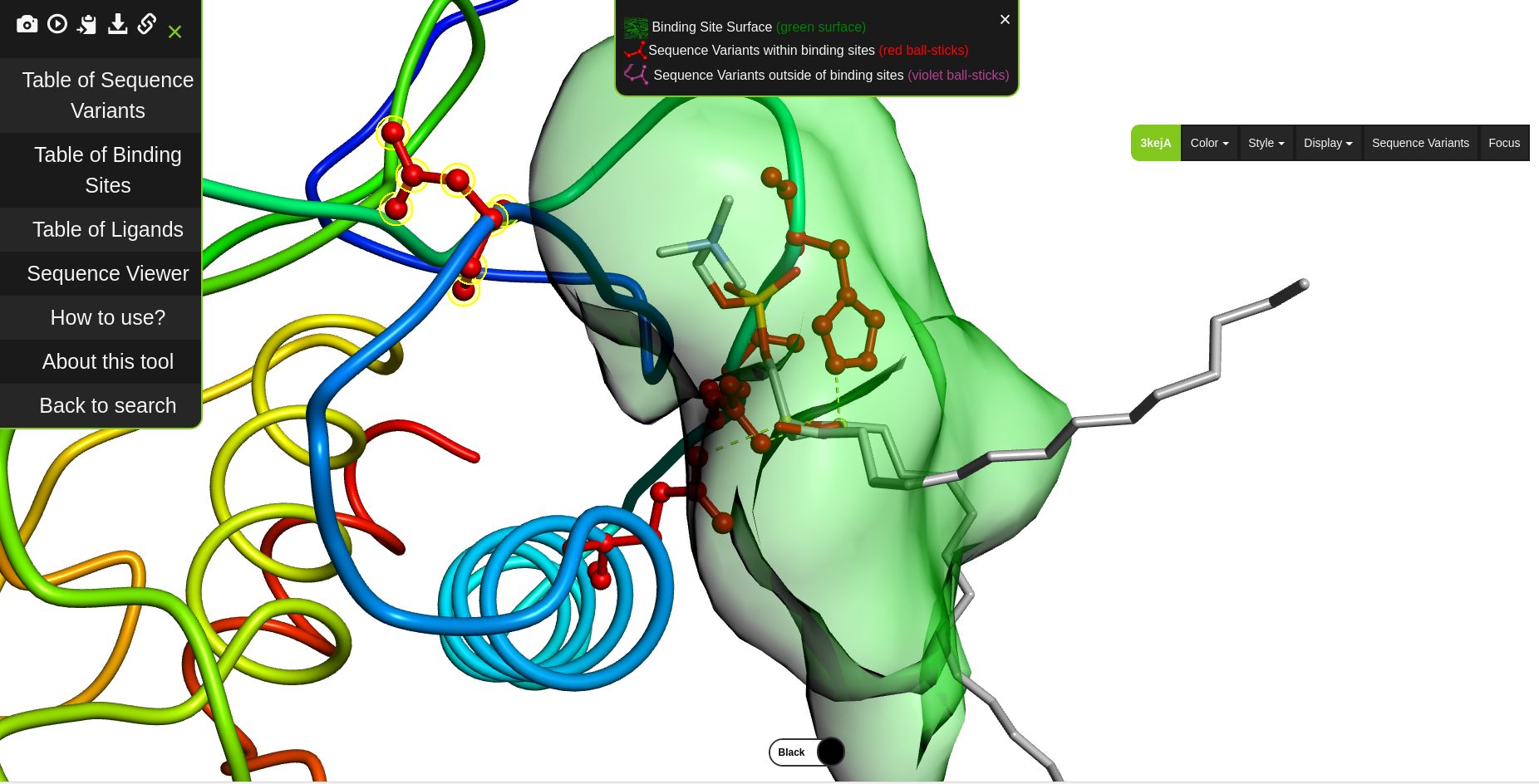
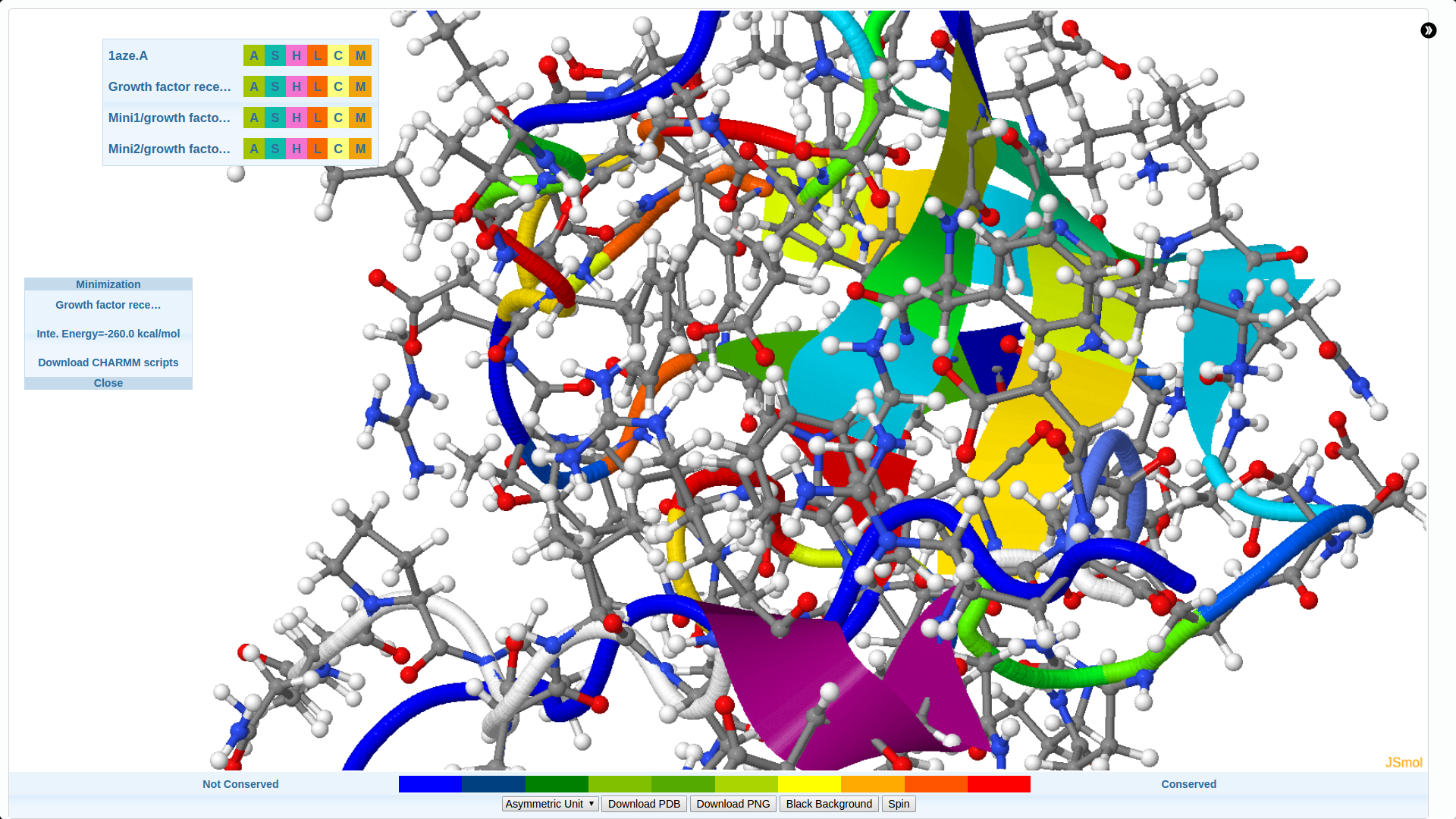
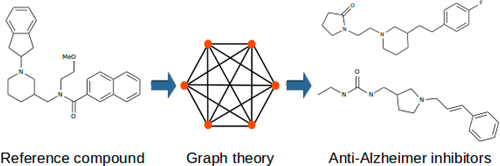
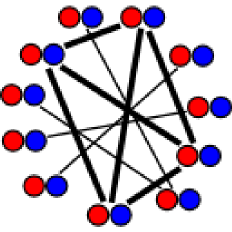
We develop new algorithms for the detection of structurally similar protein binding sites, which are based on the fact that protein surface structures are conserved in binding sites regions. These algorithms search for local similarities in physico-chemical properties in different protein surface structures independently of sequence or fold. The proteins are modeled as protein graphs, i.e., rigid 3D objects, consisting of vertices and edges. The developed algorithms are used for prediction of protein binding sites and ligand transposition in PDB protein structures.
We focus on pharmaceutically interesting proteins that could become targets for the next generation of pharmaceuticals.
We collaborate with various experimental laboratories and pharmaceutical companies. We perform experiments in silico, to eliminate as many as possible real experiments. We are not intimidated by challenging projects that involve a lot of programming, or cutting edge industrial projects. We are highly professional, experienced team of scientists and programmers that strives for highly professional attitude, and who above all love great software.