ProBiS Plugin

ProBiS plugin for PyMOL and UCSF Chimera molecular visualization programs allows prediction of binding sites and their corresponding ligands for a given protein structure. The plugin is connected via the internet to a newly prepared database of pre-calculated binding site comparisons to allow fast prediction of binding sites in existing proteins from the Protein Data Bank. The plugin enables viewing of predicted binding sites and ligands poses in three-dimensional graphics.
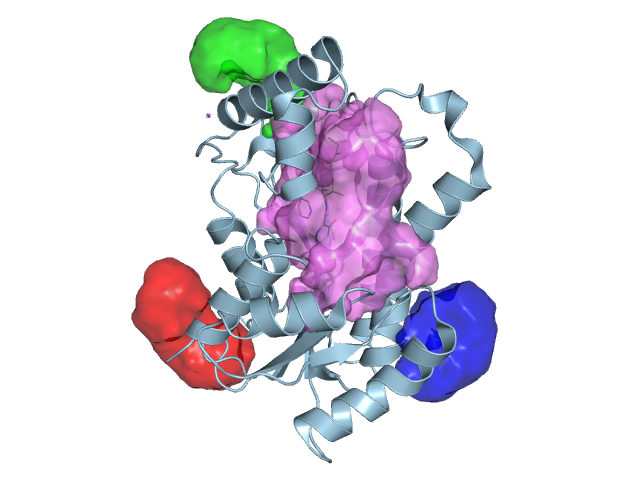
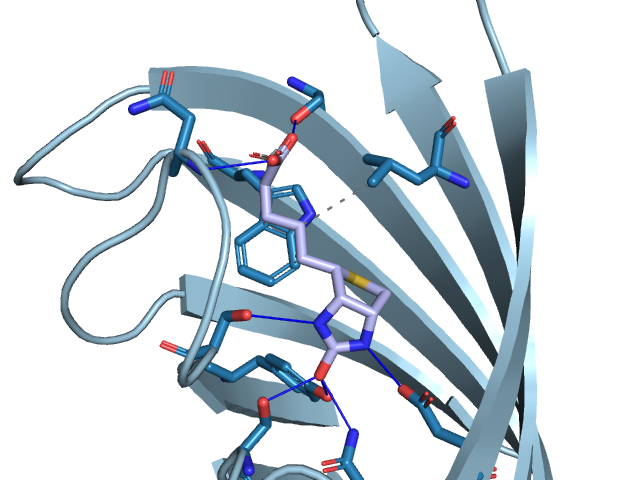
ProBiS Plugin (current version)
- ProBiS plugin for PyMOL: probis.py
- ProBiS plugin for UCSF Chimera: probis.zip
- ProBiS plugin on GitLab (Source Code): gitlab
Binding Sites Databases
Installation
Download the correct version of ProBiS plugin and follow the instructions in PyMOL or UCSF Chimera installation guide.System Requirements
The ProBiS plugin has been developed in Linux with PyMOL version 1.7.0.0 and UCSF Chimera version 1.11. In Windows it has been tested on PyMOL version 1.7.6.0 and UCSF Chimera version 1.10.2.Important Notes
- First start of the plugin can take a long time, since all the modules need to be downloaded. For complete installation of the plugin, a stable INTERNET connection is required.
- Sometimes on the first start of the plugin you may get SSL timeout error. This happens due to slow GitLab server response, and is fixed by restarting the plugin a couple of times. If the issue still persists, retry when the GitLab website is available: https://gitlab.com/tanjastular/probis-plugin.
Related Publications
Stular, Tanja, et al. "Discovery of Mycobacterium tuberculosis InhA Inhibitors by Binding Sites Comparison and Ligands Prediction", Journal of Medicinal Chemistry 59.24 (2016): 11069-11078.
Stular, T., Lesnik, S., Rozman, K., Schink, J., Zdouc, M., Ghysels, A., Liu, F., Aldrich, C. C., Haupt, V. J., Salentin, S., Daminelli, S., Schroeder, M., Langer, T., Gobec, S., Janezic, D. & Konc, J. (2016) Discovery of Mycobacterium tuberculosis InhA Inhibitors by Binding Sites Comparison and Ligands Prediction. Journal of Medicinal Chemistry, 59(24), 11069-11078.
Stular, Tanja, et al. Discovery of Mycobacterium tuberculosis InhA Inhibitors by Binding Sites Comparison and Ligands Prediction. Journal of Medicinal Chemistry, 2016, 59.24, 11069-11078.
@article{stular2016discovery,
title={Discovery of Mycobacterium tuberculosis InhA Inhibitors by Binding Sites Comparison and Ligands Prediction},
author={Stular, Tanja and Lesnik, Samo and Rozman, Kaja and Schink, Julia and Zdouc, Mitja and Ghysels, An and Liu, Feng and Aldrich, Courtney C. and Haupt, V. Joachim and Salentin, Sebastian and Daminelli, Simone and Schroeder, Michael and Langer, Thierry and Gobec, Stansilav and Janezic, Dusanka and Konc, Janez},
journal={Journal of Medicinal Chemistry},
volume={24},
number={59},
year={2016},
publisher={ACS Publications}
}
title={Discovery of Mycobacterium tuberculosis InhA Inhibitors by Binding Sites Comparison and Ligands Prediction},
author={Stular, Tanja and Lesnik, Samo and Rozman, Kaja and Schink, Julia and Zdouc, Mitja and Ghysels, An and Liu, Feng and Aldrich, Courtney C. and Haupt, V. Joachim and Salentin, Sebastian and Daminelli, Simone and Schroeder, Michael and Langer, Thierry and Gobec, Stansilav and Janezic, Dusanka and Konc, Janez},
journal={Journal of Medicinal Chemistry},
volume={24},
number={59},
year={2016},
publisher={ACS Publications}
}
%0 Journal Article
%T Discovery of Mycobacterium tuberculosis InhA Inhibitors by Binding Sites Comparison and Ligands Prediction
%A Stular, Tanja
%A Lesnik, Samo
%A Rozman, Kaja
%A Schink, Julia
%A Zdouc, Mitja
%A Ghysels, An
%A Liu, Feng
%A Aldrich, Courtney C.
%A Haupt, V. Joachim
%A Salentin, Sebastian
%A Daminelli, Simone
%A Schroeder, Michael
%A Langer, Thierry
%A Gobec, Stansilav
%A Janezic, Dusanka
%A Konc, Janez
%J Journal of Medicinal Chemistry
%V 24
%N 59
%D 2016
%I ACS Publications
%T Discovery of Mycobacterium tuberculosis InhA Inhibitors by Binding Sites Comparison and Ligands Prediction
%A Stular, Tanja
%A Lesnik, Samo
%A Rozman, Kaja
%A Schink, Julia
%A Zdouc, Mitja
%A Ghysels, An
%A Liu, Feng
%A Aldrich, Courtney C.
%A Haupt, V. Joachim
%A Salentin, Sebastian
%A Daminelli, Simone
%A Schroeder, Michael
%A Langer, Thierry
%A Gobec, Stansilav
%A Janezic, Dusanka
%A Konc, Janez
%J Journal of Medicinal Chemistry
%V 24
%N 59
%D 2016
%I ACS Publications