LiSiCA Plugin
LiSiCA PyMOL plugin is ready "out-of-box" in most Linux and Windows operating systems.
LiSiCA PyMOL Plugin (latest update Jun/13/2021)
- LiSiCA plugin v1.1.2 for PyMOL: lisica.py
- Whole package (see Manual Installation): lisica-plugin.zip
Example Input Files
- Reference molecule: reference.mol2
- Target molecules: database.mol2
Versions and Release Notes
1.1.2
- Fixed bugs: plugin size, sorting of hits by rank, other GUI changes
- Works on Linux (tested on Ubuntu 20.04) and Windows (tested on Windows 10)
- Known issue: in PyMOL 2.5.0 the visual alignment of hits is not that nice, but results are identical as in other versions of PyMOL.
1.1.1
- The plugin has been rewritten in Python 3 using Qt5 GUI
- Works with the new PyMol 2.3 and above
- This version is still available here: lisica-plugin-v111.zip (see Manual Installation)
1.0.2
- This version works for older PyMols (versions 1.x), it is still available here: lisica-plugin-v102.zip (see Manual Installation)
- Reading of gzipped files (.mol2.gz) for target molecules
- Works without internet connection
- Background color stays as set by the user
Automatic Installation (for tricks when things get wrong see Important Notes at the end of this page)
- Download lisica.py file.
-
Open PyMOL and go to Plugin → Plugin Manager.
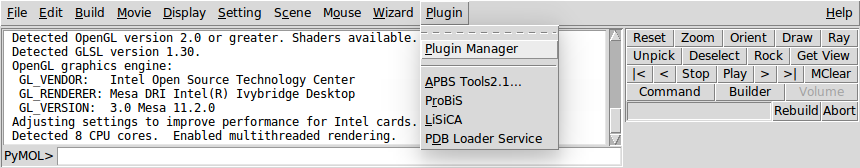
-
Select Install New Plugin category and click on the Choose file... button.
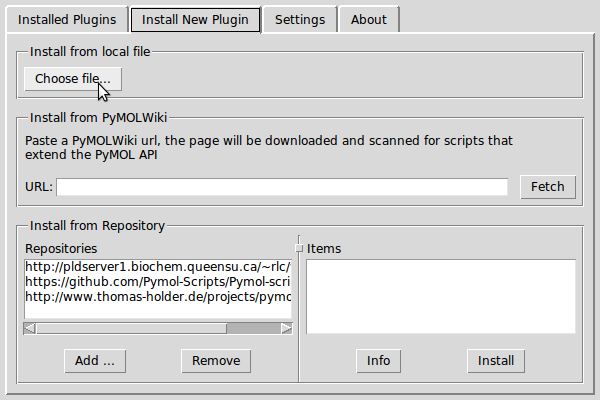
- Add the lisica.py file. In the select plugin directory window select directory and click OK
-
Close the Plugin Manager window. LiSiCA plugin should appear under Plugin → LiSiCA
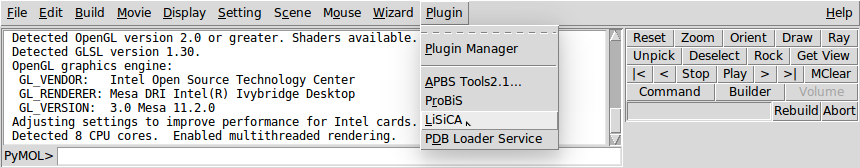
- When LiSiCA plugin is started for the first time it downloads additional required files from our server. Be patient, this may take a while.
Manual Installation (optional)
If automatic installation fails for any reason (for instance our server might be unavailable), manual installation can be accomplished like this:
- Download lisica-plugin.zip file, which contains the entire package.
- Extract it and find within the ".lisicagui" directory (the dot in front of the name means it is a hidden directory).
- Copy .lisicagui directory to your home directory.
- Install the lisica.py (see above Automatic Installation)
Important Notes
- In recent Python 3.12, eg. on Ubuntu 24.04, some libraries are deprecated. You might get "imp" and/or "distutils" missing errors. To fix these problems do "sudo apt install python3-zombie-imp python3-setuptools".
- For complete installation of the plugin, a stable internet connection is required. If a "ssh timeout" error occures, it could mean that our server is slow to respond. If this happens, retry installation a couple times.
- If installation fails with the "pmg_tk" error or similar, one or both of the tkinter and ttk libraries are not installed. To install them open the terminal and type as root "yum install tkinter python-pip" and "pip install pyttk" (in CentOS) or "apt-get install tkinter python-pip" and then "pip install pyttk" (in Ubuntu).
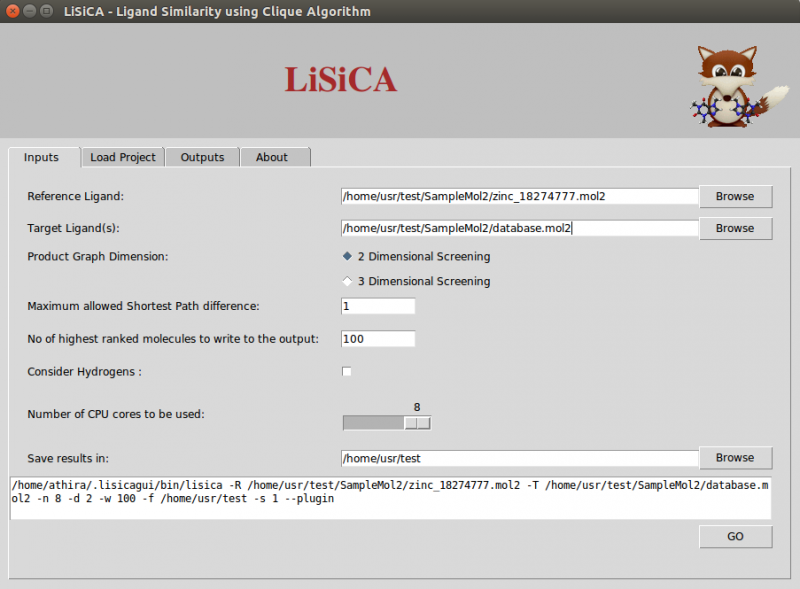
Input Tabs
- Input molecules
Both the reference and the target compound files need to be in the Tripos mol2 format. The reference file should contain only one (reference) compound; the target file may contain many compounds. If the reference file contains more than one compound, the first molecule is used as a reference. The molecules in the target file having the same name are considered as different conformers of the same molecule. By default only the best-scoring (by Tanimoto coefficient) conformer will be shown in the final output for the 3D screening option.
In the mol2 input files, the name of the molecule (ZINC ID) must be specified under the line for the "@MOLECULE" tag. For example: LiSiCA checks similarities based on the mol2 atom types. Hence SYBYL atom types have to be specified in the mol2 files. For example, under the @@
MOLECULE ZINC73655097 46 48 0 0 0 SMALL USER_CHARGES @ ATOM ... ATOM tag, each atom specification should include the SYBYL atom type, as in: In the above line from a mol2 file, the SYBYL atom type is specified in bold in column 5.1 C1 -0.0647 1.4496 -0.0592 C.3 1 <0> -0.167
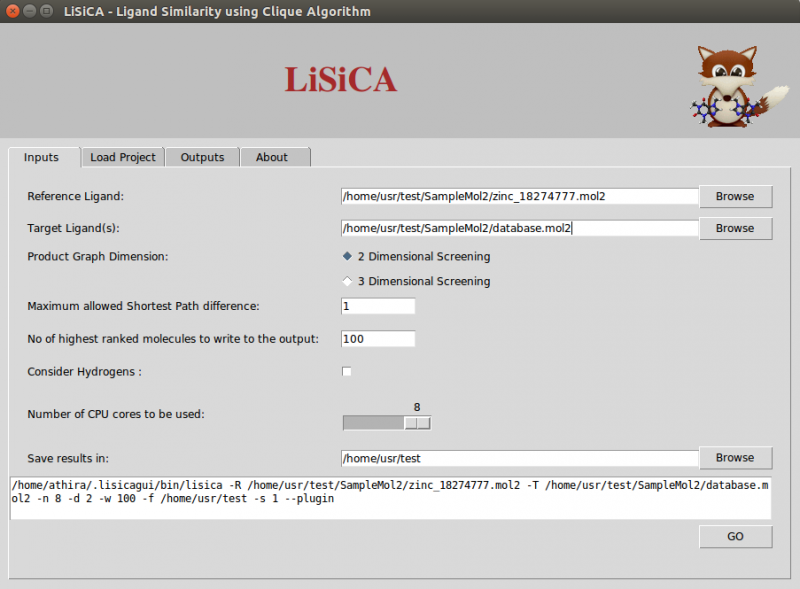
- Product graph dimensions
In 2D screening, the option Maximum Allowed Shortest path Difference corresponds to the maximum allowed difference in shortest-path length between atoms of the two compared product graph vertices. Lesser values correspond to a more rigorous screening. By default this value is a unit bond.
In 3D screening, the option Maximum Allowed Spatial Distance Difference corresponds to the maximum allowed difference in distances between atoms of the two compared product graph vertices. Lesser values correspond to a more rigorous screening. By default this value is 1 Å. The Number of Conformations option corresponds to the maximum number of outputted files of one molecule in different conformations and is to be used only for 3D screening.
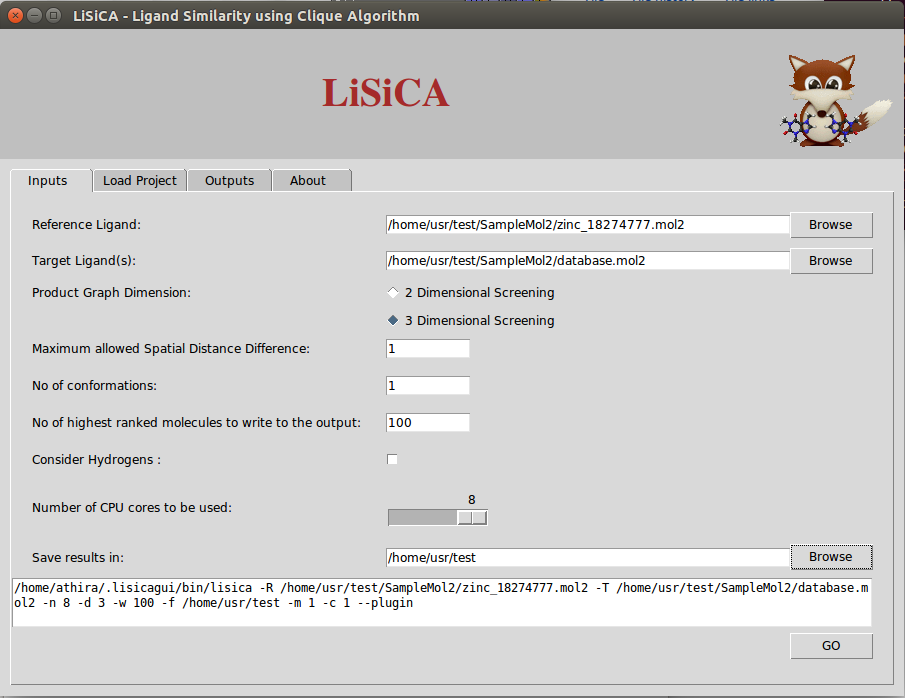
- Output
According to the value of Number of highest ranked molecules to be written to the output (say W), LiSiCA will create mol2 files of that many (W) highest scoring target molecules with a comment section at the end of the file where the matching atom pairs are displayed. This value is by default 100. The resulting mol2 files are written into a time-stamp directory in the folder specified in Save results in:. By default this folder is the user's home directory. Also, a text file named lisica_results.txt with the target molecules with (in the descending order of) Tanimoto coefficients is written to the time-stamp folder. - Other settings
The Consider Hydrogen options lets the user to choose if the hydrogen atoms are to be considered for the calculation of the similarity using the maximum clique algorithm. By default, hydrogen atoms are not considered in finding the largest substructure common to the reference and target molecules, so as to obtain the results faster.
The Number of CPU cores to be used allows for the selection of CPU threads used by LiSiCA. By default, it tries to detect the number of CPUs available.
Load Project Tab
The plugin also has a feature to load saved results. On the Load Project tab, the user can choose the directory with the saved results (containing mol2 files of each target and the reference) and the lisica_results.txt file. When the load button is clicked, the results will be loaded onto the output tab and the PyMOL Viewer window.
Output Tab
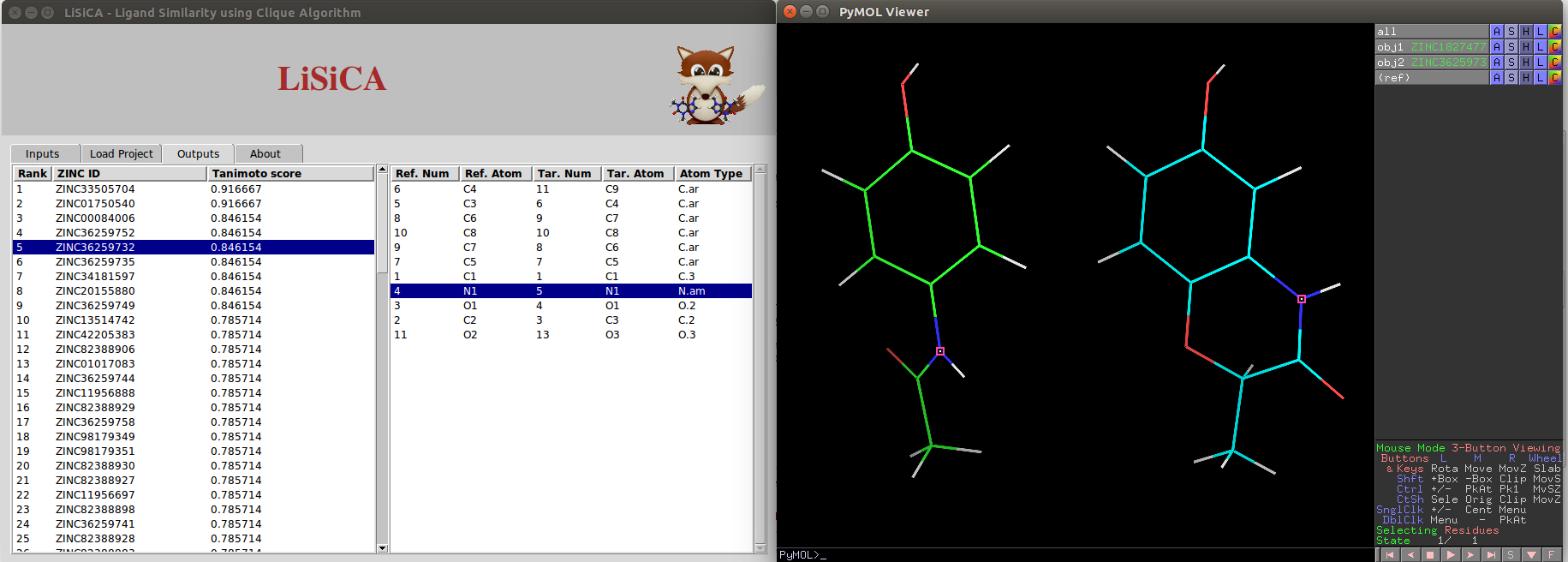
In the output tab, there are two listboxes. One contains ZINC IDs and Tanimoto Coefficients of target molecules in the decreasing order of the Tanimoto coefficient values. Any single target molecule can be selected on this listbox using a mouse click or using up/down arrow keys. The selected target molecule is displayed with the reference molecule on the PyMOL viewer window. Depending on the target molecule chosen on the first listbox, the corresponding atoms from reference and the target molecules are displayed on the other listbox. Any single pair of corresponding atoms can be selected on this listbox using a mouse click or using up/down arrow keys. The selected pair is highlighted on the PyMOL viewer window.
For 2D Screening, the two molecules (the reference and the selected target) are visualized side by side on the PyMOL viewer screen:
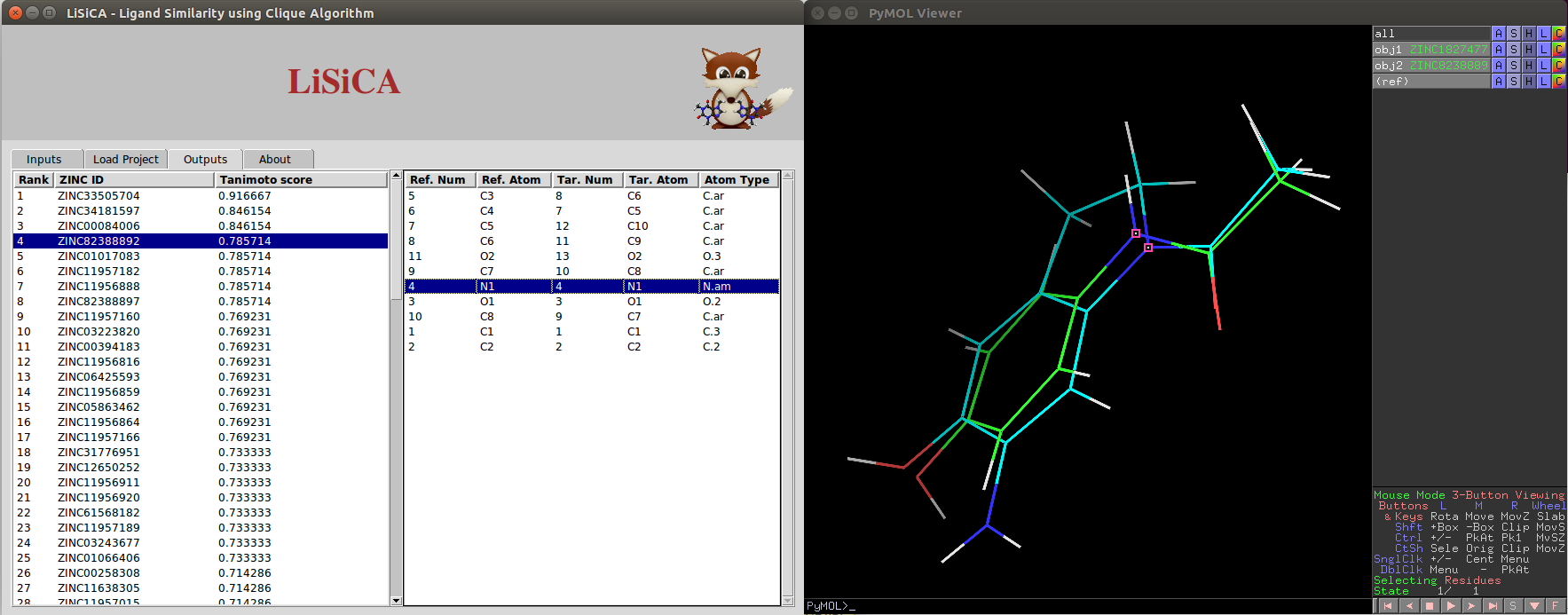
For 3D Screening, the 3D structures of the two molecules (the reference and the selected target) are superimposed using the PyMOL's pair matching function enabling easy visualisation of their similarities in the PyMOL viewer screen.
For 2D Screening, the two molecules (the reference and the selected target) are visualized side by side on the PyMOL viewer screen:

For 3D Screening, the 3D structures of the two molecules (the reference and the selected target) are superimposed using the PyMOL's pair matching function enabling easy visualisation of their similarities in the PyMOL viewer screen.

About Tab
The users can get information on new updates if available on the About tab.
Related Publications
Dilip, Athira, et al. "Ligand-based virtual screening interface between PyMOL and LiSiCA", Journal of Cheminformatics 8:46 (2016).
Dilip, A., Lešnik, S., Štular, T., Janežič, D. & Konc, J. (2016). Ligand-based virtual screening interface between PyMOL and LiSiCA. Journal of Cheminformatics, 8:46.
Dilip, Athira, et al. Ligand-based virtual screening interface between PyMOL and LiSiCA. Journal of Cheminformatics, 2016, 8:46.
@article{dilip2016lisica,
title={Ligand-based virtual screening interface between PyMOL and LiSiCA},
author={Dilip, Athira and Lešnik, Samo and Štular, Tanja and Janežič, Dušanka and Konc, Janez},
journal={Journal of Cheminformatics},
volume={8},
number={46},
year={2016},
publisher={Springer Science+Business Media}
}
title={Ligand-based virtual screening interface between PyMOL and LiSiCA},
author={Dilip, Athira and Lešnik, Samo and Štular, Tanja and Janežič, Dušanka and Konc, Janez},
journal={Journal of Cheminformatics},
volume={8},
number={46},
year={2016},
publisher={Springer Science+Business Media}
}
%0 Journal Article
%T Ligand-based virtual screening interface between PyMOL and LiSiCA
%A Dilip, Athira
%A Lešnik, Samo
%A Štular, Tanja
%A Janežič, Dušanka
%A Konc, Janez
%J Journal of Cheminformatics
%V 8
%N 46
%D 2016
%I Springer Science+Business Media
%T Ligand-based virtual screening interface between PyMOL and LiSiCA
%A Dilip, Athira
%A Lešnik, Samo
%A Štular, Tanja
%A Janežič, Dušanka
%A Konc, Janez
%J Journal of Cheminformatics
%V 8
%N 46
%D 2016
%I Springer Science+Business Media
TY - JOUR
T1 - Ligand-based virtual screening interface between PyMOL and LiSiCA
A1 - Dilip, Athira
A1 - Lešnik, Samo
A1 - Štular, Tanja
A1 - Janežič, Dušanka
A1 - Konc, Janez
JO - Journal of Cheminformatics
VL - 8
IS - 56
Y1 - 2016
PB - Springer Science+Business Media
ER -
T1 - Ligand-based virtual screening interface between PyMOL and LiSiCA
A1 - Dilip, Athira
A1 - Lešnik, Samo
A1 - Štular, Tanja
A1 - Janežič, Dušanka
A1 - Konc, Janez
JO - Journal of Cheminformatics
VL - 8
IS - 56
Y1 - 2016
PB - Springer Science+Business Media
ER -
Lešnik, Samo, et al. "LiSiCA: A Software for Ligand-Based Virtual Screening and Its Application for the Discovery of Butyrylcholinesterase Inhibitors." Journal of chemical information and modeling 55.8 (2015): 1521-1528.
Lešnik, S., Štular, T., Brus, B., Knez, D., Gobec, S., Janežič, D., & Konc, J. (2015). LiSiCA: A Software for Ligand-Based Virtual Screening and Its Application for the Discovery of Butyrylcholinesterase Inhibitors. Journal of chemical information and modeling, 55(8), 1521-1528.
LEŠNIK, Samo, et al. LiSiCA: A Software for Ligand-Based Virtual Screening and Its Application for the Discovery of Butyrylcholinesterase Inhibitors. Journal of chemical information and modeling, 2015, 55.8: 1521-1528.
@article{lešnik2015lisica,
title={LiSiCA: A Software for Ligand-Based Virtual Screening and Its Application for the Discovery of Butyrylcholinesterase Inhibitors},
author={Lešnik, Samo and Štular, Tanja and Brus, Boris and Knez, Damijan and Gobec, Stanislav and Janežič, Dušanka and Konc, Janez},
journal={Journal of chemical information and modeling},
volume={55},
number={8},
pages={1521--1528},
year={2015},
publisher={ACS Publications}
}
title={LiSiCA: A Software for Ligand-Based Virtual Screening and Its Application for the Discovery of Butyrylcholinesterase Inhibitors},
author={Lešnik, Samo and Štular, Tanja and Brus, Boris and Knez, Damijan and Gobec, Stanislav and Janežič, Dušanka and Konc, Janez},
journal={Journal of chemical information and modeling},
volume={55},
number={8},
pages={1521--1528},
year={2015},
publisher={ACS Publications}
}
%0 Journal Article
%T LiSiCA: A Software for Ligand-Based Virtual Screening and Its Application for the Discovery of Butyrylcholinesterase Inhibitors
%A Lešnik, Samo
%A Štular, Tanja
%A Brus, Boris
%A Knez, Damijan
%A Gobec, Stanislav
%A Janežič, Dušanka
%A Konc, Janez
%J Journal of chemical information and modeling
%V 55
%N 8
%P 1521-1528
%@ 1549-9596
%D 2015
%I ACS Publications
%T LiSiCA: A Software for Ligand-Based Virtual Screening and Its Application for the Discovery of Butyrylcholinesterase Inhibitors
%A Lešnik, Samo
%A Štular, Tanja
%A Brus, Boris
%A Knez, Damijan
%A Gobec, Stanislav
%A Janežič, Dušanka
%A Konc, Janez
%J Journal of chemical information and modeling
%V 55
%N 8
%P 1521-1528
%@ 1549-9596
%D 2015
%I ACS Publications
TY - JOUR
T1 - LiSiCA: A Software for Ligand-Based Virtual Screening and Its Application for the Discovery of Butyrylcholinesterase Inhibitors
A1 - Lešnik, Samo
A1 - Štular, Tanja
A1 - Brus, Boris
A1 - Knez, Damijan
A1 - Gobec, Stanislav
A1 - Janežič, Dušanka
A1 - Konc, Janez
JO - Journal of chemical information and modeling
VL - 55
IS - 8
SP - 1521
EP - 1528
SN - 1549-9596
Y1 - 2015
PB - ACS Publications
ER -
T1 - LiSiCA: A Software for Ligand-Based Virtual Screening and Its Application for the Discovery of Butyrylcholinesterase Inhibitors
A1 - Lešnik, Samo
A1 - Štular, Tanja
A1 - Brus, Boris
A1 - Knez, Damijan
A1 - Gobec, Stanislav
A1 - Janežič, Dušanka
A1 - Konc, Janez
JO - Journal of chemical information and modeling
VL - 55
IS - 8
SP - 1521
EP - 1528
SN - 1549-9596
Y1 - 2015
PB - ACS Publications
ER -